Abstract
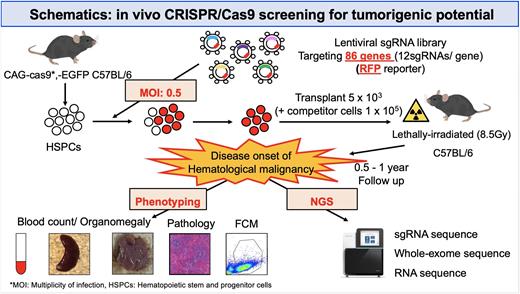
Large-scale genetic studies utilizing next-generation sequencing have identified many recurrently altered genes and revealed their overall picture in lymphoid malignancies. Although limited genes mutated at high frequency have been extensively investigated using genetically engineered mouse models, less attention has been directed to hundreds of rarely mutated genes (so-called long tail driver genes). Here we performed in vivo CRISPR loss-of-function screening to examine oncogenicity of somatic alterations, cooperability between them, and association between genotype and lineage in lymphoid malignancies. As many driver candidates mutated with various frequencies have been reported in diffuse large B-cell lymphomas (DLBCL), we constructed a custom lentiviral CRISPR library targeting 86 genes (12 sgRNAs each) in which loss-of-function alterations (mutations and deletions) are recurrently (> 5%) observed in DLBCL. These sgRNA-targeted genes also covered the majority of driver candidates of other precursor and mature lymphoid neoplasms, such as 71% and 87% of those observed in B-acute lymphoblastic leukemia (B-ALL/LBL) and peripheral T-cell lymphoma, not otherwise specified (PTCL, NOS).
Transplantation of the CRISPR library-transduced murine hematopoietic stem/progenitor (Lin-c-Kit+Sca-1+) cells caused a variety of hematologic malignancies in 180 (90%) out of 199 recipient mice, resulting in earlier death (with a median of 228 [range, 37‒476] days) of transplanted mice than control mice. Hematologic, immunophenotypic, and pathologic analyses of 132 tumors revealed that these malignancies consisted of B- and T-cell lymphomas (17% and 20%), B- and T-ALL/LBL (14% and 15%) and myeloid malignancy (14%) according to Bethesda criteria, although the diagnosis of 22 (17%) tumors could not be determined. sgRNA sequencing of 109 tumors showed that at least 1 sgRNA-targeted genes were detected in almost all (>99%) tumors, with a median of 3 (range, 1‒10) genes per tumor. Enrichment analysis of sgRNA-targeted genes demonstrated significant overrepresentation of 13 genes, such as Trp53 and Irf2bp2, pointing to stronger oncogenic capacity of these driver genes. These sgRNA-targeted genes showed lineage and tissue specificity, such as Pax5, Kmt2d, Atm, and S1pr2 for B-cell lineage and Zc3h12a and Socs1 for T-cell lineage, while only weak enrichment was observed in myeloid neoplasms. These tumors showed characteristic disease phenotype according to sgRNA-targeted genes: Pax5-targeted tumors were usually B-ALL/LBL, whereas Kmt2d-targeted tumors developed as B-cell lymphomas with earlier onset than those harboring other targeted genes. All of Zc3h12a-targeted tumors were T-cell lymphoma with CD4+CD8- immunophenotype and low clonality, while Socs1-targeted tumors primarily developed as T-ALL/LBL with CD4+CD8+ immunophenotype and frequent thymus involvement.
Whole-exome sequencing (n = 108) not only confirmed targeted disruption by sgRNAs but also identified additional driver alterations in 76% and 40% of B-cell and T-cell neoplasms, respectively, suggesting the necessity of second-hit alterations in these tumors. These additional driver alterations were associated with a specific lineage. Frequent C-terminal truncating mutations of Notch1 and loss-of-function mutations of Ikzf1 were found in T-cell malignancies, while Jak1 and Jak3 hotspot missense mutations and Flt3 internal tandem duplications were common in B-cell malignancies, suggesting cooperative relationship between specific alterations and their association with lineage. In addition, hierarchical clustering of RNA-sequencing data (n = 105) identified several clusters associated with disease phenotypes and sgRNA-targeted genes. These clusters showed distinct expression signatures, including germinal center B-cell-like signatures in Kmt2d-targeted tumors, MYC target signatures in Pax5-targeted tumors, and inflammation-related signature in Zc3h12a-targeted tumors.
In summary, our in vivo CRISPR loss-of-function screening enables high-throughput evaluation of the tumorigenic potential of many genetic abnormalities, their cooperability and association with lineage in lymphoid malignancies. Such approach accelerates understanding of genotype-phenotype association and discovery of potential therapeutic targets.
Disclosures
Koya:TOMY DIGITAL BIOLOGY CO.,LTD.: Honoraria; Scrum Inc.: Honoraria; Eisai Co., Ltd.: Honoraria. Kogure:Daiichi Sankyo Co., Ltd.: Honoraria; Nippon Shinyaku Co., Ltd.: Honoraria; Kyowa Kirin Co., Ltd.: Honoraria; Takeda Pharmaceutical Co., Ltd.: Honoraria. Ohshima:Chugai Co., Ltd.: Honoraria, Research Funding; Kyowa Kirin Co., Ltd.: Research Funding; Bristol-Myers Squibb Co.: Research Funding; Daiichi Sankyo Co., Ltd.: Research Funding. Kataoka:AstraZeneca: Honoraria; Chugai Pharmaceutical: Honoraria, Research Funding; Novartis: Honoraria; Celgene: Honoraria; Eisai: Honoraria, Research Funding; Bristol-Myers Squibb: Honoraria; Ono Pharmaceutical: Honoraria, Research Funding; Otsuka Pharmaceutical: Honoraria, Research Funding; Pfizer: Honoraria; SymBio Pharmaceuticals: Honoraria; Astellas Pharma: Honoraria; Asahi Kasei Pharma: Research Funding; Chordia Therapeutics: Research Funding; JCR Pharmaceuticals: Research Funding; Mochida Pharmaceutical: Research Funding; Teijin Pharma: Research Funding; Japan Blood Products Organization: Research Funding; Shionogi: Research Funding; Asahi Genomics: Current equity holder in private company; Genetic alterations as a biomarker in T-cell lymphomas licensed to Kyoto University and a patent for PD-L1 abnormalities as a predictive biomarker for immune checkpoint blockade therapy licensed to Kyoto University.: Patents & Royalties; Alexion Pharmaceuticals: Honoraria; Daiichi Sankyo: Honoraria; Nippon Shinyaku: Honoraria; Takeda Pharmaceutical: Honoraria, Research Funding; Janssen Pharmaceutical: Honoraria; Kyowa Kirin: Honoraria, Research Funding; Sumitomo Dainippon Pharma: Honoraria, Research Funding; AbbVie: Honoraria; Meiji Seika Pharma: Honoraria; Sanofi: Honoraria; Keio University School of Medicine: Other: NA; National Cancer Center Research Institute: Other: NA.
Author notes
Asterisk with author names denotes non-ASH members.